
SKU : G3468-25000U
หมวดหมู่ : Biochemicals , 1. Chemical and Reagents , Buffers , Servicebio ,
แบรนด์ : Servicebio
Share
Product Information
Product Name | Product Number | Specification |
Peptide-N-Glycosidase F (PNGase F) | G3468-25000U | 25000 U |
Product Introduction
Peptide-N-Glycosidase F , referred to as PNGase F, is derived from the glycosidase F of Chryseobacterium miricola . It is recombinantly expressed by Escherichia coli and can cleave high-mannose, hybrid and complex oligosaccharide glycoproteins linked by asparagine. The cleavage site of PNGase F is the amide bond between the N-acetylglucosamine (GlcNAc) on the glycoprotein side and the asparagine residue, and at the same time, the aspartyl on the enzymatically hydrolyzed protein is converted to aspartic acid. It is mainly used to remove N-glycosylation of proteins.
Source: Derived from Chryseobacterium miricola , recombinantly expressed in Escherichia coli;
Enzyme activity definition: The amount of enzyme required to remove more than 95% of carbohydrates from 10 μg of denatured RNase B at 37°C in 1 h is defined as one unit of enzyme activity;
Purity and concentration: Purity detected by SDS-PAGE>95%; residual endogenous nucleic acid <1 pg/μL (qPCR detection); 500 U/μL;
Inactivation or inhibition: Heating at 75°C for 10 min will inactivate;
Enzyme storage buffer: 20 mM Tris-HCl, 50 mM NaCl, 5 mM EDTA, 50% Glycerol, pH 7.5;
10х Denaturing Buffer: 5% SDS, 400 mM DTT;
10х Native Buffer: 400 mM sodium phosphate, pH 7.5;
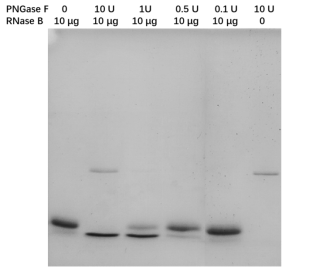
Figure 1. Deglycosylation effect of substrate RNase B treated with PNGase F. After denaturation treatment of substrate RNase B, 10 μg of the substrate was added with different amounts of PNGase F (0, 10, 1, 0.5, 0.1 U), incubated at 37°C for 1 h, and then all were detected by PAGE electrophoresis.
Storage and transportation
Transported with wet ice; stored at -20, valid for 12 months.
composition
Component Number | Component | G 3468 |
G3468-1 | Peptide-N-Glycosidase F (PNGase F) | 50 μL |
G3468-2 | 10х Denaturing Buffer | 1 mL |
G3468-3 | 10х Native Buffer | 1 mL |
manual | 1 serving | |
Procedure
1. Protein deglycosylation under denaturing conditions:
Add 1 μL 10х Denaturing Buffer and dH 2 O to 10 μL reaction system in 1-20 μg protein sample , and incubate the system at 100 for 10 min. Then add 2 μL 10х Native Buffer, 2 μL 10% NP-40, 1 μL PNGase F and appropriate amount of dH 2 O to 20 μL reaction system, and incubate at 37 for 1 h. (Note: The activity of PNGase F will be inhibited by SDS, so NP-40 must be added under denaturing conditions)
2. Protein deglycosylation under non-denaturing conditions:
Add 2 μL 10х Native Buffer, 2-5 μL PNGase F and appropriate amount of dH 2 O to 1-20 μg protein sample to 20 μL reaction system and incubate at 37 for 4-24 h. (Protein deglycosylation under non-denaturing conditions requires longer reaction time or more enzymes)
Precautions
1. The activity of PNGase F will be inhibited by SDS, so NP-40 must be added under denaturing conditions.
2. The simplest way to assess the extent of protein deglycosylation is by SDS-PAGE gel electrophoresis.
3. The enzyme should be placed in an ice box and stored at -20 immediately after use.
4. For your safety and health, please wear a lab coat and disposable gloves when operating.
The product is for scientific research purposes only and not for clinical diagnosis!