
SKU :
Option name 1
Categories : Biochemicals , 1. Chemical and Reagents , Buffers , Servicebio ,
Brand : Servicebio
Share
Product Information
Product Name | Cat. No. | Spec. |
2×In-Fusion Cloning Mix Plus | G3351-20T | 20 T |
G3351-100T | 100 T |
Product Description/Introduction
The 2×In-Fusion Cloning Mix Plus offer increased cloning efficiency over previous generations of In-Fusion kits (G3350,2×In-Fusion Cloning Mix), especially for multiple fragments. It is designed for fast, directional cloning of one or 2~5 multiple fragments of DNA into any vector. It fuses DNA fragments (e.g., PCR-generated inserts and linearized vectors) efficiently and precisely by recognizing 15-25 bp overlaps at their ends. These 15-25 bp overlaps can be engineered by designing primers for amplification of the desired sequences.The simultaneous insertion of multiple fragments greatly simplifies the experimental steps, improves cloning efficiency and saves your time.
1.Single-fragment primer design: 15-25 bp sequences consistent with both ends of the linearized vector were introduced into the 5' end of the amplification primer of the insert. Design the diagram below:
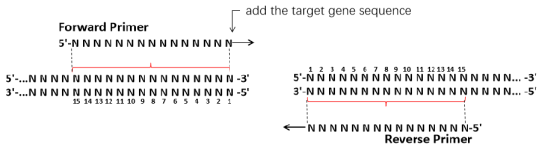
2.Multi-fragment primer design: The principles of primer design at both ends of the vector are the same as those of single-fragment primers. Reset zone between fragment primer design principle is as follows: There is a 15-25 bp overlap between the reverse primer of Fragment 1 and the forward primer of Fragment 2. The reverse primer for Fragment 1 includes the overlapping region and the reverse specific primer region, the forward primer for Fragment 2 includes the overlapping region and the forward specific primer region, and so on (overlapping regions are shown in red). To improve efficiency, the overlapping regions between fragments can be increased and their Tm values are guaranteed to be consistent. The design is as follows:

Storage and Shipping Conditions
Ship with wet ice; stored at -20°C, valid for 12 months.
Product Contents
Component | G3351-20T | G3351-100T |
2×In-Fusion Cloning Mix Plus | 100 μL | 5 x 100 μL |
pUC19 (Linearized, Control Vector, 5 ng/μL) | 10 μL | 10 μL |
Control Inset (10 ng/μL) | 10 μL | 10 μL |
Manual | One copy | |
Assay Protocol / Procedures
Perform ligation reaction
1. To an autoclaved, 1.5-ml microcentrifuge tube, add the following(recommend 10-uL reaction system):
Component | Volume |
2×In-Fusion Cloning Mix Plus | 5 μL |
Vector DNA | X μL |
Insert DNA | Y μL |
Nuclease-Free Water | Add to 10 μL |
2. Mix gently and centrifuge briefly to bring the contents to the bottom of the tube.
3. For single-fragment recombination reactions, incubate at 50°C for 15 minutes; For the multi-fragment recombination, incubate at 50°C for 30 minutes.
NOTE: For 3-5 multi-fragment recombination, increase the reaction time will yield more colonies, but should not exceed 1 h).
4. Place the tube on ice and Proceed immediately to Perform transformation reaction.
Perform transformation reaction
5. Add appropriate ligation reaction into Chemically Competent E. coli (such as DH5α, Top10, etc.)and mix gently. Do not mix by pipetting up and down.
6. Incubate for 30 minutes on ice.
7. Heat-shock the cells for 30 seconds in a 42°C water bath.
8. Immediately place the tubes on ice and incubate for 2 minutes.
9. Add 900 µL of room temperature SOC or LB medium. Cap the tube tightly and shake the tube horizontally at 225 rpm for 1 hour at 37°C.
10. Spread required volume of transformation reaction on a prewarmed LB plate containing corresponding antibiotics.
11. Incubate plates overnight at 37°C.
Analyze transformants
Pick an individual colony from the transformation plate, analyze transformants by colony PCR or restriction enzyme digestion.
Note
1. The vector DNA and insert DNA should be gel purified and analyse their quality and concentration by electrophoresis. Water can be omitted in ligation reaction if the concentration is low.
2. The Tm value between the overlapping regions of multiple fragments should be consistent and >60.
3. It is recommended that the molar ratio of vector and insert is 1:1~1:3; when 2-3 fragments are connected, the molar ratio between each fragment is 1:1, and the ligation reaction system can be scaled up in equal proportions.
4. If the total volume of vector and insert is more than 5 μL, you may scale the ligation system to 20 μL.
5. The volume of the ligation product should not exceed 1/10 of the volume of the competent cells, otherwise the transformation efficiency will be significantly reduced. The volume of the ligation product and the competent cells can be increased in equal proportions (for example, 20 μL of ligation system transforms 200 μL of competent cells).
6. The 2×Universal Ligation Mix shoule be kept at -20°C until within 5-10 minutes of use and returned immediately to -20°C after use. It is recommended to freeze in aliquots to reduce freeze-thaw cycles.
7. If electroporation is used for transformation, vector DNA and insert DNA should be purified by column method or ethanol precipitation method.
Different types of plasmid construct experimental process diagram
1. The recombinant plasmid construct experimental process diagram
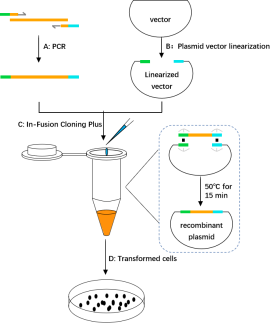
A. Insert DNA fragments were obtained by PCR amplification:The 15-25 bp homologous sequence (green and light blue) from the linearized vector end was introduced into the 'end of the forward/reverse primers of the target fragment, and the 5' and 3' end sequences of the amplified product were completely consistent with the two end sequences of the linearized vector, respectively.
B. Plasmid vector linearization:Restriction endonuclease or reverse PCR were used to obtain linearized vectors.
C. In-Fusion Cloning Plus: The linearized carrier and the purified and recovered insert fragment were mixed in proportion, and the reaction was completed at 50 for 15 min.
D. Transformed cells:Reaction products directly into competent escherichia coli cells, monoclonal colony growth pick it out from the tablet for PCR, enzyme digestion and sequencing experimental screening positive clones.
2. Single point mutations recombinant plasmid construct experimental process diagram
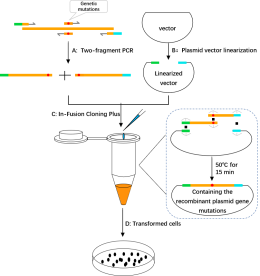
A. Two-fragment PCR(PCR amplifies DNA fragments at both ends of the mutation site) : Complementary primers were designed at the mutation site (orange), and primers were designed at both ends of the mutant gene to introduce homologous sequences at the end of the 15-25 bp linearized vector (green and light blue). Using different primers for the PCR amplification, respectively, mutations in two pieces with the homologous sequences of two insert fragment (include) gene mutations, respectively.
B. Plasmid vector linearization:Restriction endonuclease or reverse PCR were used to obtain linearized vectors.
C. In-Fusion Cloning Plus: The linearized carrier and the purified and recovered insert fragment were mixed in proportion, and the reaction was completed at 50 for 15 min.
D. Transformed cells:Reaction products directly into competent escherichia coli cells, monoclonal colony growth pick it out from the tablet for PCR, enzyme digestion and sequencing experimental screening positive clones.
3. Schematic diagram of the experimental process of multi-fragment insertion recombinant plasmid construction
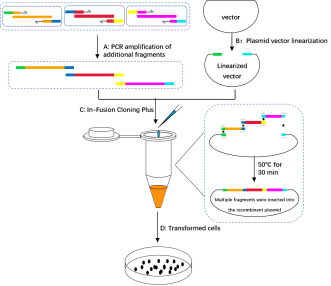
A. PCR amplification of additional fragments: The homologous sequences (marked green, dark blue, yellow and light blue) were added to the 5' end of the amplification primers, so that there was a 15-25 bp homologous sequence between the amplification products and between the amplification products and the linearized vector. Different primer pairs were used to amplify the DNA fragments (marked orange, red and purple).
B. Plasmid vector linearization:Restriction endonuclease or reverse PCR were used to obtain linearized vectors.
C. In-Fusion Cloning Plus: The linearized carrier and the purified and recovered insert fragment were mixed in proportion, and the reaction was completed at 50 for 15 min.
D. Transformed cells:Reaction products directly into competent escherichia coli cells, monoclonal colony growth pick it out from the tablet for PCR, enzyme digestion and sequencing experimental screening positive clones.
For Research Use Only!